Genomic basis of parallel serpentine adaptation
Genomic basis of parallel serpentine adaptation

Our finding demonstrates that large pool of pre-existing variation is the major source of adaptive alleles in autotetraploids. However, rapid selection on de novo mutations is still feasible, altogether indicating broad evolutionary flexibility of lineages with doubled genomes. You can discover more here. It was a nice highly collaborative project between our lab (Veronika, Jakub, Majda and Doubravka), Levi Yant's and David Salt's labs (U Nottingham) and Christian Parisod's lab (U Bern).
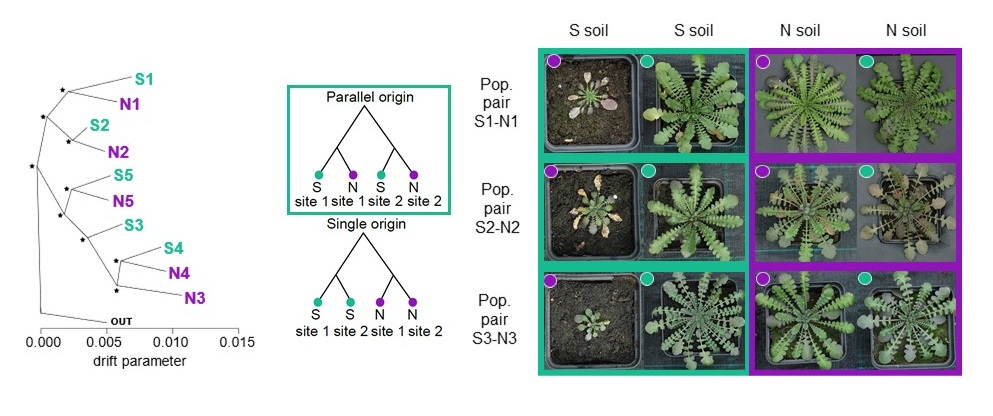
Leveraging such fivefold naturally replicated design, we found a substantial genomic parallelism in serpentine adaptation. Then, we modelled parallel selection and infer dominant role of sampling from a shared pool of alleles across the variable tetraploid populations.

However, we also discovered an exceptional locus with parallel de novo mutations! It encodes a central transporter TPC1 protein, shown to act as a global hub mediating stress signalling, with two distinct serpentine-specific mutations in otherwise conserved site at calcium selectivity gate.
